F. Gräter: Mechano-sensing mechanisms in the desmosome from molecular simulations

Principal investigator
Prof. Dr. Frauke Gräter
Heidelberg Institute for Theoretical Studies
Schloß-Wolfsbrunnenweg 35
69118 Heidelberg
Tel: +49 (0)6221-533267 (office)
frauke.graeter(at)h-its.org

SPP funded collaborator
Dr. Csaba Daday
Heidelberg Institute for Theoretical Studies
Schloß-Wolfsbrunnenweg 35
69118 Heidelberg
csaba.daday(at)h-its.org
SPP funded collaborator
Dr. Fan Jin
Heidelberg Institute for Theoretical Studies
Schloß-Wolfsbrunnenweg 35
69118 Heidelberg
Tel: +49 (0)6221–533–254 (office)
fan.jin(at)h-its.org
Summary
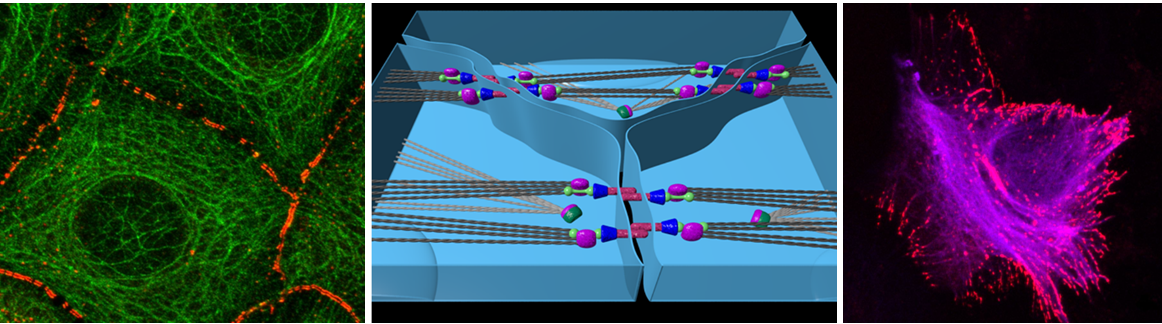
Desmosomes integrate mechanical stress into biochemical networks at the cell-cell interface. How desmosomal proteins respond to mechanical force such that their structure and function is altered is currently unknown. Our objective is to characterize the mechanical properties and putative force-sensing function of a major desmosomal component, desmoplakin. The spectrin-repeat fragment of desmoplakin features an SH3 domain with a peculiar and cryptic binding site and currently unknown function. The spectrin-SH3 interaction is a hot spot for mutations involved in skin and cardiac diseases, underlining its pivotal role in desmoplakin function. We will put two putative roles of the SH3 domain, and thereby of desmoplakin, to test, namely a mechanically stabilizing function and a mechano-sensing function, which might not exclude each other. To this end, we will perform Molecular Dynamics simulations to monitor the spectrin/SH3 fragment and larger constructs of desmoplakin under tensile forces.
The simulations will allow us to quantify the extent to which the SH3 domain, when being subjected to mechanical force, can stabilize the spectrin repeats against unfolding and/or can expose its binding site for partners involved in downstream chemical signalling. We will also examine desmoplakin variants, lacking the SH3 domain or carrying disease mutants, to shed further light on desmoplakin’s force-carrying and force-sensitive role in stressed desmosomes.
To test eventual redox regulation of desmoplakin’s force response, we will subject the protein at different oxidation states to a newly developed disulfide swapping algorithm and monitor variations in the unfolding mechanism. Our results, after validation by single molecule experiments to be performed at King’s College London, can help to interpret and guide future in vitro and in vivo experiments. We expect our work to reveal, for the first time, a direct role of desmoplakin in force transmission and conversion of mechanical stress into biochemical signals.
Expertise
- computational biophysics
- Molecular Dynamics simulations
- coarse grained models and Brownian dynamics
- bioinformatics
- quantum chemistry